- HOME
- Members
- Principal Investigators
- Principal Investigators
- Hiroki Kurihara
RESEARCH
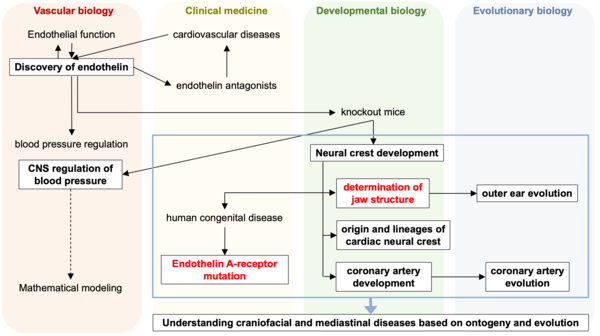
1. Craniofacial development
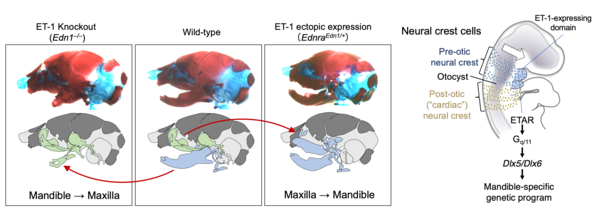
The branchial (pharyngeal) arches are a segmental series of bulging structures common and characteristic for all vertebrate embryos. They are mainly formed by migratory cranial neural crests, which give rise to various skeletal components including the jaw and middle ear structures. We have revealed that endothelin-1 (ET-1), first identified as an endothelium-derived vasoconstrictor peptide, and its receptor ETAR signaling acts as a molecular switch that determines the lower jaw identity by using mouse genetics.
PI's main research track. Highlights in red are shown in the figures below.
From this research, several new insights in the field of evolutionary developmental biology have been obtained. In mouse embryos, the absence of the Endothelin signal results in the failure to form the eardrum and the external ear canal. In contrast, in chicken embryos, there is instead an occurrence of duplication formation. This reveals that the eardrum is a product of convergent evolution that independently evolved in mammals and reptiles/birds. Furthermore, the facial structure of mammals deviates significantly from the common facial development pattern found in jawed vertebrates. It undergoes dynamic evolution, altering the tip of the upper jaw in reptiles and amphibians to create a protruding nose while simultaneously developing new bones to reshape the mouth tip. This has been elucidated through experimental embryology and comparative studies, including the analysis of fossils.
Endothelin signaling determines the identity of jaw structures.
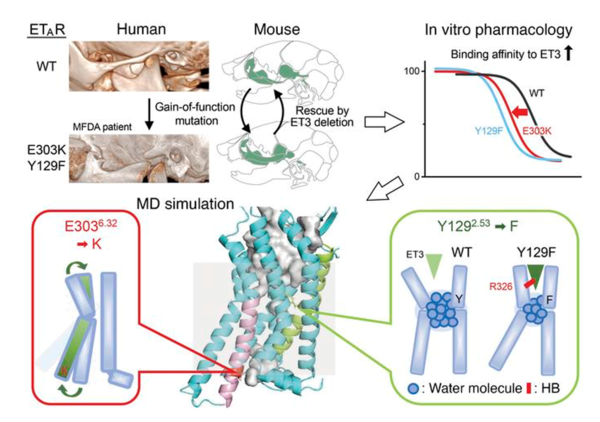
Recently, two different recurrent mutations in the Endothelin-A receptor (ETAR) gene EDNRA have been identified as causative for mandibulofacial dysostosis with alopecia (MFDA), a rare congenital disorder in humans. The mechanism of its pathogenesis has been elucidated through the use of mouse genetics, pharmacological experiments, and molecular dynamics (MD) simulations. This has revealed that the single amino acid EDNRA mutations distant from the ligand binding site allosterically affect ligand binding, inhibiting ligand dissociation and increasing binding affinity.
Mandibulofacial dysostosis with alopecia results from ETAR gain-of-function mutations via allosteric effects on ligand binding. (from https://www.jci.org/articles/view/151536)
2. Cardiac development
We have advanced our analysis regarding three cell types that migrate through the cardiac outflow tract and contribute to heart formation:
1) We clarified that mesodermal cells from the extraembryonic somatopleure, which serves as the amniotic primordium, migrate into the embryo and differentiate into cardiomyocytes and vascular endothelial cells. We also revealed that BMP and FGF signals are involved in inducing these differentiations.
2) Through transcriptome analysis of neural crest cells migrating into the heart and spatial transcriptome analysis, we unveiled a diverse cell population, including undifferentiated cell groups resembling stem cells and precursors, as well as smooth muscle-like cells. We also conducted a time-evolving lineage prediction for these cell populations.
3) In coronary artery formation, we found the involvement of lymphatic endothelial cells that are believed to originate from the periaortic capillary plexus around the aortic root during embryonic development. We demonstrated the role of Semaphorin signals in this process.
Furthermore, through comparative anatomical and developmental analysis of coronary arteries, we elucidated that in amphibians, teleost fish, and cartilaginous fish, the primary vessels formed during embryonic development directly supply the heart, whereas, in amniotes (mammals, birds, reptiles), these primary vessels undergo remodeling during development, forming a new entrance to establish the coronary arteries. This provides a new foundation for understanding the causes of congenital coronary artery abnormalities.
3. Angiogenesis
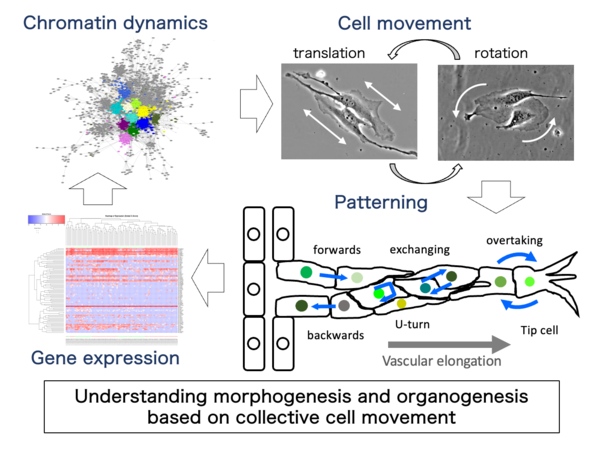
Angiogenesis is a morphogenetic process that produces branching vascular structures during embryogenesis and various (patho-)physiological conditions. We have identified characteristic cellular behaviors in angiogenic processes, including dynamic changes in forward-backward movement, tip cell overtaking and resultant cell mixing. Although the cellular behaviors appear complex and arbitrary, different types of mathematical modeling (stochastic vs. deterministic) and experimental verification indicated that some deterministic cell-cell interactions are critical for vascular elongation and possibly branching.
Recently, we found differences in branch-forming capacity among cell types and some regularities in directional cell movement using in vitro angiogenesis experiments using mouse vascular explants and an endothelial cell line by refined cell-tracking system. Together with single-cell analyses of cell movement and gene expression, novel mathematical modeling and experimental verification using constitutional approaches are under way in collaboration with Professor Tetsuji Tokihiro (Graduate School of Mathematical Sciences, The University of Tokyo, Musashino University) and his colleagues, to elucidate the possible cellular mechanisms underlying branch formation in angiogenesis.
To elucidate the collective motility characteristics of endothelial cells that enable sprout elongation, we analyzed the dynamics of mouse pancreatic islet microvascular-derived endothelial cell line MS-1 cells. We discovered that their directional movement is influenced by cell contacts, with a combination of linear motion with directionality and rotational movement as its basis. Through various inhibitors and genetic modifications using genome editing, it has become evident that these endothelial cell-specific motility characteristics are specifically regulated by adhesion molecules such as VE-cadherin. Currently, we are advancing the elucidation of the coordination mechanism between cell motility and transcriptional control by contrasting gene expression profiles and cell behavioral phenotypes.