- HOME
- News & Events
- Publications
- 【Publications】Application of targeted enrichment to next-generation sequencing of retroviruses integ...
Publications
【Publications】Application of targeted enrichment to next-generation sequencing of retroviruses integrated into the host human genome
June 22 2016
Yorifumi Satou
Paper information
Paola Miyazato, Hiroo Katsuya, Asami Fukuda, Yoshikazu Uchiyama, Misaki Matsuo, Michiyo Tokunaga, Shinjiro Hino, Mitsuyoshi Nakao & Yorifumi Satou*
Application of targeted enrichment to next-generation sequencing of retroviruses integrated into the host human genome (* corresponding author)
Scientific Reports 6, Article number: 28324 (2016) doi:10.1038/srep28324
Highlights
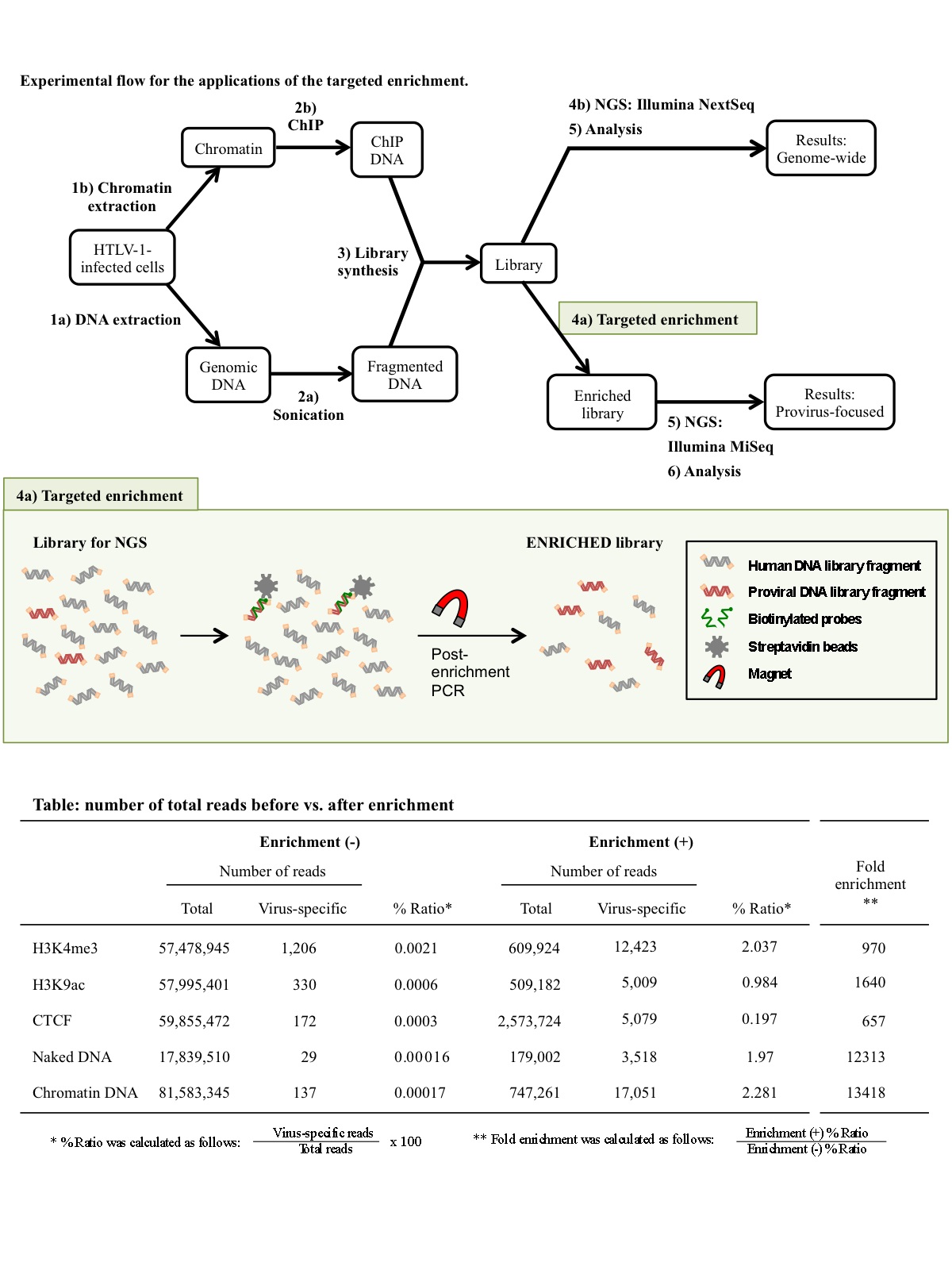
*Proviruses, integrated forms of retroviruses, are currently the focus of attention due to the difficulty encountered in their eradication from infected people.
*We report a novel probe-based hybridization capture method that facilitates the analysis of integrated proviruses, with high through-put depth.
*The method versatile enough to be applied to the analysis of DNA and RNA sequences and its flexibility tolerates a certain degree of genetic variations in the analyzed sequences.
Abstract
The recent development and advancement of next-generation sequencing (NGS) technologies have enabled the characterization of the human genome at extremely high resolution. In the retrovirology field, NGS technologies have been applied to integration-site analysis and deep sequencing of viral genomes in combination with PCR amplification using virus-specific primers. Nevertheless, viral sequences are poorly detected, because proviral DNA is very scarce compared to the human genomic DNA. Here, we have developed and evaluated the use of biotinylated DNA probes for the capture of viral genetic fragments from a library prepared for NGS. Our results demonstrated that viral sequence detection was hundreds or thousands of times more sensitive after enrichment, enabling us to reduce the economic burden that arises when attempting to analyze the epigenetic landscape of proviruses by NGS. Taken together, we propose that this approach is a powerful tool to clarify the mechanisms of transcriptional and epigenetic regulation of retroviral proviruses that have, until now, remained elusive.